class: center, middle, inverse, title-slide # Meta-analysis course: part 1 : Systematic reviews, meta-analysis, and introduction to R ### Thomas Pollet (<span class="citation">@tvpollet</span>), Northumbria University ### 2019-09-16 | <a href="http://tvpollet.github.io/disclaimer">disclaimer</a> --- ## Outline for this section. * What is a systematic review / meta-analysis? * Baby steps in RStudio / R. --- ## Sources used. Can be found at end of slides. I relied heavily on Harrer et al. (2019) and Weiss and Daikler (2016). Among others I found this book very interesting... . <img src="Koricheva.jpg" width="275px" style="display: block; margin: auto;" /> --- ## GoSoapbox Go to [www.gosoapbox.com](www.gosoapbox.com) Enter code: 257883396 --- ## Tell me about yourself. **Thomas switches on questions** <img src="https://media.giphy.com/media/26nfqZcvPsRwZFImQ/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## Need for synthesis. * Textbook examples. Problematic. -- What is a seminal single study in your field? <img src="https://media.giphy.com/media/QWA9p190VpNLxO1ryW/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## Synthesis --> Synthesis. Types of synthesis: * Narrative review. * Vote counting. * Combining probabilities. * Systematic reviews: Qualitative and Quantitative (contains a Meta-analysis) --- ## Narrative reviews. Generally invited contributions by experts (e.g., _TREE, Phil. Trans. B., Annual Review of X_) Some quite narrow in scope, some quite comprehensive and large. Covered papers could range from dozens to hundreds. Useful for perspectives, historical development refining concepts. But by no means complete ('systematic'). --- ## Drawbacks of narrative reviews. Traditional narrative reviews are biased: - Convenience sample - Inefficient handling of large and complex information (variation in outcome measure). At best a large table. - Hard to reconstruct. Roberts et al. (2006) analysed 73 reviews in the area of conservation management, only 30% reported which sources were consulted for the review (N=73). Thus, likely reflects reviewer bias,... . - Typical focus on dichotomous statistical significance testing (blinded by _p_ values). Effect found or not. (Publication bias) <img src="https://media.giphy.com/media/QWA9p190VpNLxO1ryW/giphy.gif" width="250px" style="display: block; margin: auto;" /> --- ## Vote counting. In its simplest form, 3 categories : Significant + / Non-significant / Significant - . Alternative forms: linear + , linear -, curvilinear (different shapes possible), ... , no effect. -- Advantage: simple/broadly applicable. -- Disadvantages: - How to handle variability in outcomes? - one vote counts the same for N=10 vs. N=1000 - no information on magnitude of effect - low power for small effect - statistical power decreases as more studies are added (?!, Hedges & Olkin (1980)) --> Formal systematic reviews and meta-analysis are better. --- ## Combining probabilities. Exists since Fisher (1946). Quite popular in social sciences. Basically tallying _p_ values. Most common is summing across a normal distribution <div class="figure" style="text-align: center"> <img src="Youngronaldfisher2.jpg" alt="Ronald Fisher (1913)" width="375px" /> <p class="caption">Ronald Fisher (1913)</p> </div> --- ## Combining probabilities: Strengths and weaknesses A plus is that it is broadly applicable (widespread use of _p_ values). -- Solves some of the issues with vote counting: _p_ = .04 and _p_ = .06 . -- However, as with vote counting not very informative. -- Small _p_ value large effect (but uncertain?) or large sample size (with small effect size) -- Too liberal, if many tests and one low _p_ nearly always significant (Cooper, 2010:160) --- ## Systematic review [Cochrane](https://consumers.cochrane.org/what-systematic-review): _A systematic review summarises the results of available carefully designed healthcare studies (controlled trials) and provides a high level of evidence on the effectiveness of healthcare interventions. Judgments may be made about the evidence and inform recommendations for healthcare_ Systematic, structured and objective. Documentation of all research steps (literature retrieval, data entry, coding, etc.) and relevant decisions <img src="https://media.giphy.com/media/8FiRcf3XYx2i4/giphy.gif" width="250px" style="display: block; margin: auto;" /> --- ## Comparison of methods <div class="figure" style="text-align: center"> <img src="Table1.1_Koricheva.png" alt="Comparison of methods by Koricheva et al. 2013" width="625px" /> <p class="caption">Comparison of methods by Koricheva et al. 2013</p> </div> --- ## Systematic reviews. Most useful when: * there is a substantive research question. * several empirical studies have been published. (Sometimes a mini-) * there is uncertainty about the results. Does not always contain a meta-analysis. --- ## Definitions (from Petticrew & Roberts) **Systematic (literature) review** "A review that strives to comprehensively identify, appraise, and synthesize all the relevant studies on a given topic. Systematic reviews are often used to test just a single hypothesis, or a series of related hypothesis." **Meta-analysis** "A review that uses a specific statistical technique for synthesizing the results of several studies into a single quantitative estimate (i.e. a summary effect size)" ([Petticrew & Roberts, 2006](https://books.google.co.uk/books?id=ZwZ1_xU3E80C&lpg=PR5&ots=wYT6sNGQMu&dq=petticrew%20roberts&lr&pg=PR5#v=onepage&q=petticrew%20roberts&f=false)). --- ## What is the relationship between a systematic review and meta-analysis * Remember qualitative systematic reviews also exist. * The term (quantitative) research synthesis or (quantitative) systematic review denotes the entire research process, which has qualitative as well as quantitative elements. * The quantitative part of a (quantitative) systematic review is called a meta-analysis. * You don't need a systematic review in order to have a meta-analysis (but recommended) --> example mini-meta-analysis within a paper (Cumming et al., 2014; [Goh et al. 2016](https://www.northeastern.edu/socialinteractionlab/wp-content/uploads/2016/10/goh.etal_.2016.SPPC_.pdf)) <img src="https://media.giphy.com/media/l4FGw4d101Sa0pGTe/giphy.gif" width="250px" style="display: block; margin: auto;" /> --- ## Purpose... . What is a systematic review (/meta-analysis) for? * Describe the existence (or prevalence) of phenomenon (e.g., [prevalence of PTSD](https://tvpollet.github.io/pdfs/Morina_et_al_2018.pdf), Morina et al. (2018)) * Exploration: explore a research question (e.g., compile a list of risks for job loss) * Formally evaluate hypotheses (e.g., "the challenge hypothesis" ([Wingfield et al., 1990](https://www.reed.edu/biology/professors/srenn/pages/teaching/2008_syllabus/2008_readings/9_wingfield_etal_1990.pdf)) - [Varying androgen responses to mating, breeding or territorial behaviour in avian males?](https://www.sciencedirect.com/science/article/pii/S0003347205003623) * to formally evaluate (medical, social, educational) interventions. For example, [stereotype threat and performance in girls](https://daneshyari.com/article/preview/363552.pdf) --- ## What is a meta-analysis? Karl Popper (1959): _"Non-reproducible single occurrences are of no significance to science."_ Gene Glass (1976, p.3): _"An analysis of analyses, I use it to refer to the statistical analysis of a large collection on analysis results from individual studies for the purpose of integrating the findings"_ -- Studies addressing a common research question are 'synthesized'. Synthesizing: * describing the quality of the sample (e.g., in terms of selection bias). * calculating an overall outcome statistic (e.g, Pearson _r_, odds ratio) * determining and describing the amount of heterogeneity in the outcome statistics. * trying to explain the above heterogeneity by means of, for example, meta-regression. -- In sum: Meta-analysis --> A class of statistical techniques. --- ## Superiority of meta-analysis. Research synthesis is superior for summarising results: * Outcomes of many studies are reduced to few numbers; * but still accounting for potential heterogeneity of the studies. Dealing with heterogeneous study findings: * Describe amount of heterogeneity. * Study characteristics can be potential predictors to explain heterogeneity among study results (e.g., research design, sample characteristics). --- ## Evidence based movement <img src="EBMupdated.jpg" width="600px" style="display: block; margin: auto;" /> --- ## Types of meta-analysis. We can categorise based on: * Study design: experimental, quasi-experimental, observational * Individual patient data (IPD, raw data available) vs. aggregated patient data (APD, based on publications) --> IPD is preferred (e.g., able to directly assess data quality, [Cooper & Patall, 2009](https://www.ncbi.nlm.nih.gov/pubmed/19485627)) --- ## Limitations of meta-analysis. * A lot of effort if you want to do it properly. * Only as 'powerful' as the inputs. --> cannot convert correlational studies into experimental ones. * Bias in study selection is very difficult to avoid. (We can only try and estimate its extent). * Analysis of between-study variation via meta-regression is inherently correlational. <img src="limitation.jpg" width="275px" style="display: block; margin: auto;" /> --- ## Common Criticisms. * Apples and oranges: Interest lays in 'fruit salad' / Heterogeneity. * _Garbage in / Garbage out_ : Meta-analysis is nothing more than waste management? Solution: Systematically examine the quality of studies (coding) and examine differences in outcomes. * Missing data / publication bias: Affects _any_ kind of research. Meta-analysis allows for methods addressing these problems. <img src="trash.gif" width="300px" style="display: block; margin: auto;" /> --- ## Common Myths about meta-analysis (Littell et al. 2001) * Meta-analyses require a medical perspective and require experimental data on treatments (RCT) --> False. (e.g., meta-analysis on observed correlations, prevalence, etc.) * Meta-analyses require large number of studies and/or large sample sizes --> False. Sometimes just 2(!) <img src="X-files.gif" width="300px" style="display: block; margin: auto;" /> --- ## Golden standards in systematic reviews / meta-analyis. Not many do so in (Evolutionary) Psychology and related fields. * [Cochrane collaboration](https://www.cochrane.org/) in medicine. * [Campbell collaboration](https://campbellcollaboration.org/) covers policy (social sciences). Publishes Campbell Systematic Reviews (expected to be updated min. every 3 years). * [PROSPERO](https://www.crd.york.ac.uk/prospero/) register for prospective reviews relating to health ([Booth et al., 2011](https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(10)60903-8/fulltext)). Example [here](https://www.crd.york.ac.uk/prospero/display_record.php?RecordID=32695) * [PRISMA guidelines](http://prisma-statement.org/). Detailed [checklist](http://prisma-statement.org/documents/PRISMA%202009%20checklist.pdf) <img src="https://media.giphy.com/media/l0HlKB7bThU9CzTqM/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## Steps in a meta-analyis. Overview and then we'll zoom in on some steps,... . 7 steps according to Cooper (2010:12-ff): * Formulating the problem * Searching the literature * Gathering information from studies * Evaluating the quality of studies * Analyzing and integrating the outcomes of studies * Interpreting the evidence * Presenting the results (incidentally 9 according to [this paper](https://peerj.com/preprints/2978/).) --- ## Formulating the problem. **Q**: What evidence will be relevant to the key hypothesis in the meta-analysis? -- **A**: Define: - Variables of interest. - research designs - historical (everything or since X), geographical, theoretical context. --> discriminate relevant from irrelevant. -- Procedural variations can lead to relevant/irrelevant or included but tested for moderating influence. --> Example: *[Red and attractiveness](https://journals.sagepub.com/doi/full/10.1177/1474704916673841)*. --- ## PICOS **P**opulation **I**ntervention **C**omparison **O**utcome **S**tudy Type **S**ample **P**henomon of **I**nterest **D**esign **E**valuation **R**esearch type ([Methley et al. 2014](https://bmchealthservres.biomedcentral.com/articles/10.1186/s12913-014-0579-0)) --- ## Searching the literature **Q**: What procedures do we use to find the relevant literature? **A** Identify: (a) Sources (journals/databases) (b) Search terms. Again procedural variations can lead to differences between researchers. --- ## Gather information from studies **Q**: Which information from each study is relevant to the research question of interest? **A**: Collect relevant information reliably Recurring theme: Procedural variations might lead to differing conclusions between researchers due to (a) what information is gathered (b) difference in coding (especially when multiple coders) (c) deciding on independence of studies (d) specificity of data needed. --- ## Gathering information from papers. * Typically takes 9 to 24 months time to do a systematic review (estimate from [Centre for Reviews and Dissemination at York](https://www.york.ac.uk/crd/)). * Guidelines [PRISMA](http://www.equator-network.org/reporting-guidelines/prisma/) (from the EQUATOR network). * Best practice: [PRISMA checklist](http://prisma-statement.org/documents/PRISMA%202009%20checklist.pdf) --- ## Evaluating research results. **Q**: What research should be included based on the suitability of research methods used and any issues (e.g., DV not measured accurately) **A**: Identify and apply criteria on which studies should be included or not. (e.g., include only studies on conscious evaluation and not on priming) Again, procedural variations might influence which studies remain included and which are not. --- ## Analyzing and integrating the outcomes of studies. **Q**: How should we combine and summarise the research results? **A**: Decide on how to combine results across studies and how to test for substantial differences between studies. Surprise: There could be variations... (i.e., choice of effect size measure) --- ## Interpreting the evidence. **Q**: What conclusions can be drawn based on the compiled research evidence? **A**: Summarize the cumulative research evidence in temrs of strength, generality, and limitations. Variation between researchers in labelling results as important and attention to variation between studies in attention to detail. --- ## Presenting the results **Q**: What information needs to be included in the write-up of the report. **A**: Follow journal guidelines and determine what methods / results readers of the paper will have to know. ([OSF](http://osf.io) for everything.) Variation in reporting exists and could have consequences on how much other researchers trust your report and the degree to which they can reconstruct your findings. --- ## Steps in a meta-analyis. Overview and then we'll zoom in on some steps,... . 7 steps according to Cooper (2010:12-ff): * Formulating the problem * Searching the literature * **Gathering information from studies** * Evaluating the quality of studies * Analyzing and integrating the outcomes of studies * Interpreting the evidence * Presenting the results --- ## Coding. Coding scheme. Purpose: * Express study results in a standardized form * Find predictors which could explain variation in study outcomes * Anticipate "reviewer 2" and code what you need to address possible criticisms of your review. <img src="Reviewer2.jpg" width="300px" style="display: block; margin: auto;" /> --- ## What can be coded? **Thomas goes to GoSoapbox.** --- ## What can be coded? Information on: * Outcome measures (e.g., effect size) * Characteristics of study design / sample (e.g., Number of women, year of publication, ... ). * Coding process itself. <img src="https://media.giphy.com/media/3oKIPnAiaMCws8nOsE/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## Coding outcomes. Note some redundant: * Effect size(s) * Variable(s)/construct(s) represented in the effect size * Subsample information, if relevant (e.g., scores split out by men/women) * Sample size(s) (effect size specific) * Means or proportions * Standard deviations or variances * Calculation procedure (effect size specific) (how estimated? transformed?) --- ## Study descriptors. Theoretical variables: * For example on economic games. Degree of control player A has over B Methods and procedures: * Sampling procedure * Design (e.g., Sexy red effect: manipulation of clothes vs. background) * Attrition / Drop out Descriptors of paper: * Publication form (published/unpublished) * Publication year * Country of publication (WEIRD or not) * Study sponsorship * ... --- ## Reliability of coding. * Golden standard: At least 2 raters independently code. Resolve any issues via discussion. * Calculate interrater reliability (Hayes & Krippendorff, 2007; also see Yeaton & Wortman, 1993) --- ## Tools to help with coding process. * Could just use Excel / Googlesheet. * [Revman](https://community.cochrane.org/help/tools-and-software/revman-5). * [Metagear](http://lajeunesse.myweb.usf.edu/metagear/metagear_basic_vignette.html) in R. Mostly for reviewing abstracts. <img src="https://media.giphy.com/media/3oKIPqsXYcdjcBcXL2/giphy.gif" width="350px" style="display: block; margin: auto;" /> --- ## Flow Chart. Use [this](http://prisma.thetacollaborative.ca/) to do so online. <img src="PRISMA_2009_flow_diagram.png" width="300px" style="display: block; margin: auto;" /> --- ## Steps in a meta-analyis. Overview and then we'll zoom in on some steps,... . * Formulating the problem * Searching the literature * Gathering information from studies * Evaluating the quality of studies * Analyzing and integrating the outcomes of studies * Interpreting the evidence * **Presenting the results** --- ## Multiple standards for reporting. * APA , Meta-analyis reporting standards [MARS](https://wmich.edu/sites/default/files/attachments/u58/2015/MARS.pdf) -- updated [here](https://www.apa.org/images/amp-amp0000191_tcm7-228474.pdf) * [PRISMA](http://www.prisma-statement.org/) <img src="https://media.giphy.com/media/26BGGp9NoFefWZFPG/giphy.gif" width="350px" style="display: block; margin: auto;" /> --- ## Software packages. * [Revman](https://community.cochrane.org/help/tools-and-software/revman-web) software for lit. reviews. * [CMA](https://www.meta-analysis.com/) --> extensive and good support but "pay to play". Other packages also exist. * [R](https://cran.r-project.org/) --> Free, reproducible, modifiable for your purposes. [JASP](https://jasp-stats.org) relies on R and can do many of the things I'll cover... . <img src="https://media.giphy.com/media/bxYGpFWlvuWnC/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## I already know R... If you already know about R and RStudio. Need internet connection for quick install. ```r install.packages("Rcade") library("Rcade") games$Pacman games$CathTheEgg games$`2048` games$SpiderSolitaire games$Core games$CustomTetris games$GreenMahjong games$Pond games$Mariohtml5 # Doesn't work? games$BoulderDash # Doesn't work? ``` --- ## The R environment. Install [R Studio](https://www.rstudio.com/products/rstudio/download/) and [R](https://cran.r-project.org/). Runs on Windows / Linux / OSX. ```r install.packages("tidyverse") install.packages("meta") install.packages("metafor") install.packages("RISmed") ``` Thomas opens RStudio and hopes for the best! --- ## Support. * Google is your friend, ... . * [Stackoverflow](www.stackoverflow.com). * [Crawley (2013)](ftp://ftp.tuebingen.mpg.de/pub/kyb/bresciani/Crawley%20-%20The%20R%20Book.pdf) / [Wickham & Grolemund (2016)](https://r4ds.had.co.nz/) <img src="https://media.giphy.com/media/QVxeI5qhmlXAkqaAro/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## Extremely minimal introduction in R and RMarkdown. RStudio - New file. Click File New --> R markdown. --> Document---> Html. (Many other options incl. presentations) This will be the core in which you will complete your analyses. RMarkdown can be rendered in .html / .word / .pdf --- ## RMarkdown Press the knit button! <img src="samplermarkdown.png" width="700px" /> --- ## HTML Congrats. You generated a webpage! -- The bit between the ticks are R code. The text in between is [Markdown](https://www.markdownguide.org/). A very simple language. -- Occasionally .html or . latex code interspersed. -- You can make .pdf but .html is suitable for most purposes... . -- If you want to make PDFs you'll need a latex distribution. On Windows, you need Miktex, installed here in the lab. On OSX, MacTeX. On Linux, TexLive. More info [here](https://www.latex-project.org/get/). --- ## First coding ever. <img src="http://i0.kym-cdn.com/entries/icons/original/000/021/807/4d7.png" width="200px" /> Delete what's between the ticks. Enter: - Sys.Date() and Click "Run Current Chunk" Should give you: ```r Sys.Date() ``` ``` ## [1] "2019-09-16" ``` --- ## Sys.time() - Sys.time() and Click "Run Current Chunk" Should give you: ```r Sys.time() ``` ``` ## [1] "2019-09-16 18:47:37 -03" ``` --- ## R and RStudio R is not really a programme but rather works based on packages. Some basic operations can be done in base R, but mostly we will need packages. First we install some packages. This can be done via the install.packages command. In RStudio you also have a button to click. **Thomas shows Rstudio button** Try installing the 'ggplot2' package via the button. --- ## Loading a package. * packages: and then tick ggplot2 * Or: ```r library(ggplot2) #loading ggplot2 ``` '#' to write comments in your code, which does not get read. Again, if you know [Latex](https://www.latex-project.org/), you can also incorporate this (as with .html) --- ## R as a calculator. Use ; if you want several operations. ```r 2+3; 5*7; 3-5 ``` ``` ## [1] 5 ``` ``` ## [1] 35 ``` ``` ## [1] -2 ``` --- ## Mathematical functions. Mathematical functions are shown below (Crawley, 2013:17). <img src="Figure1_crawley.png" width="700px" /> --- ## Let's make a variable... We often want to store things on which we'll do the calculations. ```r thomas_age<-37 ``` **IMPORTANT** Variable names in R are case sensitive, so Thomas is not the same as thomas. Variable names should not begin with numbers (e.g. 2x) or symbols (e.g. %x or $x). Variable names should not contain blank spaces (use body_weight or body.weight not body weight). --- ## Object modes (atomic structures) **integer** whole numbers (15, 23, 8, 42, 4, 16) **numeric** real numbers (double precision: 3.14, 0.0002, 6.022E23) **character** text string (“Hello World”, “ROFLMAO”, “DR Pollet”) **logical** TRUE/FALSE or T/F --- ## Object classes **vector** object with atomic mode **factor** vector object with discrete groups (ordered/unordered) **matrix** 2-dimensional array **array** like matrices but multiple dimensions **list** vector of components **data.frame** "matrix –like" list of variables of same # of rows --> **This is the one you care most about.** Many of the errors you potentially run into have to do with objects being the wrong class. (For example, R is expecting a data.frame, but you are offering it a matrix). --- ## Assignment, or how to label a vector (or variable) **<-** assign, this is to assign a variable. At your own risk you can also use = . [Why?](http://blog.revolutionanalytics.com/2008/12/use-equals-or-arrow-for-assignment.html) **c(...)** combine / concatenate **seq(x)** generate a sequence. **[]** denotes the position of an element. --- ## Examples. ```r # Now let's do some very simple examples. seq(1:5) # print a sequence ``` ``` ## [1] 1 2 3 4 5 ``` ```r thomas_height<-188.5 # in cm thomas_height # prints the value. ``` ``` ## [1] 188.5 ``` ```r # number of coffee breaks in a week number_of_coffees_a_week<-c(1,2,0,0,1,4,5) number_of_coffees_a_week ``` ``` ## [1] 1 2 0 0 1 4 5 ``` ```r length(number_of_coffees_a_week) # how many elements ``` ``` ## [1] 7 ``` --- ## Days of the week. ```r days<-c("Mon","Tues","Wed","Thurs","Friday", "Sat", "Sun") days ``` ``` ## [1] "Mon" "Tues" "Wed" "Thurs" "Friday" "Sat" "Sun" ``` ```r days[5] # print element number 5 -- Friday ``` ``` ## [1] "Friday" ``` ```r days[c(1,2,3)] # print elements 1,2,3 ``` ``` ## [1] "Mon" "Tues" "Wed" ``` --- ## Replacing things. ```r days[5]<-"Fri" # replace Friday with Fri days ``` ``` ## [1] "Mon" "Tues" "Wed" "Thurs" "Fri" "Sat" "Sun" ``` ```r days[c(6,7)] <- rep("Party time",2) # write Sat and Sun as Party time days ``` ``` ## [1] "Mon" "Tues" "Wed" "Thurs" "Fri" ## [6] "Party time" "Party time" ``` --- ## Try it yourself (in duos) Use # to annotate your code. 1. Make an atomic vector with your height. If you don't know your metric height: 'guess'. 2. Make a vector for the months of the year. 3. Print the 6th and 9th month 4. Replace the July/August with vacation in your vector. --- ## Special Values **NULL** object of zero length, test with is.null(x) **NA** Not Available / missing value, test with is.na(x) **NaN** Not a number, test with is.nan(x) (e.g. 0/0, log(-1)) **Inf, -Inf** Positive/negative infinity, test with is.infinite(x) (e.g. 1/0) --- ## Is.numeric / etc. ```r is.numeric(thomas_age) ``` ``` ## [1] TRUE ``` ```r is.numeric(days) ``` ``` ## [1] FALSE ``` ```r is.atomic(thomas_age) ``` ``` ## [1] TRUE ``` ```r is.character(days)[1] ``` ``` ## [1] TRUE ``` --- ## Checking for missings: is.na() ```r is.na(thomas_age) ``` ``` ## [1] FALSE ``` ```r is.na(days) ``` ``` ## [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE ``` --- ## Combining vectors into a matrix. Combining vectors is easy, use c(vector1,vector2) Combining column vectors into one matrix goes as follows. **cbind()** column bind **rbind()** row bind --- ## Example with coffee data ```r coffee_data<-cbind(number_of_coffees_a_week,days) coffee_data # this is what the matrix looks like. ``` ``` ## number_of_coffees_a_week days ## [1,] "1" "Mon" ## [2,] "2" "Tues" ## [3,] "0" "Wed" ## [4,] "0" "Thurs" ## [5,] "1" "Fri" ## [6,] "4" "Party time" ## [7,] "5" "Party time" ``` ```r coffee_data<-as.data.frame(coffee_data) # make it a dataframe. is.data.frame(coffee_data) ``` ``` ## [1] TRUE ``` --- ## Try it yourself. Together with your partner: 1. combine the two vectors with your heights. (Remember the order!) (or make a new one!) 2. make a vector with your ages (in the same order as 1.) 3. make a dataframe called 'team' using cbind 4. check that it is a dataframe. --- ## Making a matrix from scratch. ```r # nr: nrow / nc; ncol matrix(data=5, nr=2, nc=2) ``` ``` ## [,1] [,2] ## [1,] 5 5 ## [2,] 5 5 ``` ```r matrix(1:8, 2, 4) ``` ``` ## [,1] [,2] [,3] [,4] ## [1,] 1 3 5 7 ## [2,] 2 4 6 8 ``` ```r as.data.frame(matrix(1:8,2,4)) ``` ``` ## V1 V2 V3 V4 ## 1 1 3 5 7 ## 2 2 4 6 8 ``` --- ## Setting a work directory. Normally you would do this at the start of your session. If you don't it'll live where your file lives. Which isn't always bad (and) So, this is where you would read and write data,... . ```r setwd("~/Dropbox/Teaching_MRes_Northumbria/Lecture1") # the tilde just abbreviates the bits before # mostly you would use setwd("C:/Documents/Rstudio/assignment1") # for windows. Dont use ~\ # Linux: setwd("/usr/thomas/mydir") ``` --- ## Writing away data. One of the most versatile formats is .csv comma separated value file (readable in MS Excel) ```r write.csv(coffee_data, file= 'coffee_data.csv') ### no row names. write.csv(coffee_data, file= 'coffee_data.csv', row.names=FALSE) ### ??write.csv to find out more ``` SPSS (install 'haven' first!) , note the **different** notation! ```r require(haven) write_sav(coffee_data, 'coffee_data.sav') ``` --- ## Read in data. If it is in the same folder. I have reloaded the 'haven' package. ```r require(haven) coffee_data_the_return<-read_sav('coffee_data.sav') ### use the same notation as with setwd to get the path ``` Even from (public) weblinks. Here in .dat format. head() shows you the first lines. ```r require(data.table) mydat <- fread('http://www.stats.ox.ac.uk/pub/datasets/csb/ch11b.dat') head(mydat) ``` ``` ## V1 V2 V3 V4 V5 ## 1: 1 307 930 36.58 0 ## 2: 2 307 940 36.73 0 ## 3: 3 307 950 36.93 0 ## 4: 4 307 1000 37.15 0 ## 5: 5 307 1010 37.23 0 ## 6: 6 307 1020 37.24 0 ``` --- ## Some basic data analyses / manipulations. This follows [Whickham & Grolemund (2017)](https://r4ds.had.co.nz/). [library](https://yihui.name/en/2014/07/library-vs-require/) (instead of require - require tries to load library). I'll switch. ```r library(nycflights13) ``` ```r library(tidyverse) ``` ``` ## ── Attaching packages ──────────────────────────────────────────────────────────── tidyverse 1.2.1 ── ``` ``` ## ✔ ggplot2 3.2.0 ✔ purrr 0.3.2 ## ✔ tibble 2.1.3 ✔ dplyr 0.8.3 ## ✔ tidyr 0.8.3 ✔ stringr 1.4.0 ## ✔ readr 1.3.1 ✔ forcats 0.4.0 ``` ``` ## ── Conflicts ─────────────────────────────────────────────────────────────── tidyverse_conflicts() ── ## ✖ dplyr::between() masks data.table::between() ## ✖ dplyr::filter() masks stats::filter() ## ✖ dplyr::first() masks data.table::first() ## ✖ dplyr::lag() masks stats::lag() ## ✖ dplyr::last() masks data.table::last() ## ✖ purrr::transpose() masks data.table::transpose() ``` --- ## Conflicts. Take careful note of the conflicts message printed loading the tidyverse. It tells you that dplyr conflicts with some functions. Some of these are from base R. If you want to use the base version of these functions after loading dplyr, you’ll need to use their full names: stats::filter() and stats::lag() <img src="http://s2.quickmeme.com/img/83/83f3188fb441ef8a727dac6715f50478910342242eb4f6e974675a3f75c044d8.jpg" width="200px" /> --- ## NYC Flights This data frame contains all 336,776 flights (**!**) that departed from New York City in 2013. From the US Bureau of Transportation Statistics, and is documented in ?flights. ```r nycflights13::flights ``` ``` ## # A tibble: 336,776 x 19 ## year month day dep_time sched_dep_time dep_delay arr_time ## <int> <int> <int> <int> <int> <dbl> <int> ## 1 2013 1 1 517 515 2 830 ## 2 2013 1 1 533 529 4 850 ## 3 2013 1 1 542 540 2 923 ## 4 2013 1 1 544 545 -1 1004 ## 5 2013 1 1 554 600 -6 812 ## 6 2013 1 1 554 558 -4 740 ## 7 2013 1 1 555 600 -5 913 ## 8 2013 1 1 557 600 -3 709 ## 9 2013 1 1 557 600 -3 838 ## 10 2013 1 1 558 600 -2 753 ## # … with 336,766 more rows, and 12 more variables: sched_arr_time <int>, ## # arr_delay <dbl>, carrier <chr>, flight <int>, tailnum <chr>, ## # origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>, hour <dbl>, ## # minute <dbl>, time_hour <dttm> ``` ```r # Lets make it available to our enivronment. flights<-(nycflights13::flights) ``` --- ## Tibbles. Tibbles are data frames. But with some tweaks to make life a little easier. You can turn a dataframe into a tibble with as_tibble() --- ## Notice anything in particular? **int** stands for integers. **dbl** stands for doubles, or real numbers. **chr** stands for character vectors, or strings. **dttm** stands for date-times (a date + a time). --- ## But I want to see everything. View() ```r View(flights) ``` <img src="https://pics.me.me/this-is-bran-bran-knows-everything-yet-he-does-not-27295932.png" width="300px" /> --- ## 'dplyr' basics. Pick observations by their values: **filter()**. Reorder the rows: **arrange()**. Pick variables by their names **select()**. Create new variables with functions of existing variables **mutate()**. Collapse many values down to a single summary **summarise()**. --- ## Data cleaning... Let's filter out some missings for departure delay (dep_delay) Here we make a _new_ dataset <img src="https://i.pinimg.com/736x/30/66/ac/3066ac69ae68ac200ace0ca8fe3882c3--friday-memes-broken-city.jpg" width="300px" /> --- ## filter() ```r # notice '!' for 'not'. flights_no_miss<-filter(flights, dep_delay!='NA') ``` <img src="https://i.imgflip.com/13ui7g.jpg" width="300px" /> --- ## Logical operations. & is “and”, | is “or”, and ! is “not” <img src="transform-logical.png" width="500px" /> --- ## = vs. == When filtering you'll need: the standard suite: >, >=, <, <=, != (not equal), and == (equal). Common mistake: = instead of == --- ## Floating point numbers floating point numbers are a problem. Computers cannot store infinite numbers of digits. ```r sqrt(3) ^ 2 == 3 ``` ``` ## [1] FALSE ``` ```r 1/98 * 98 == 1 ``` ``` ## [1] FALSE ``` <img src="floating_cat.jpg" width="175px" /> --- ## Solution: near() ```r near(sqrt(3) ^ 2, 3) ``` ``` ## [1] TRUE ``` ```r near(1/98*98, 1) ``` ``` ## [1] TRUE ``` --- ## Basic statistics. Let's look at the delays with departure (dep_delay). Note the dollar sign ($) for selecting the column ```r mean(flights_no_miss$dep_delay) ``` ``` ## [1] 12.63907 ``` ```r median(flights_no_miss$dep_delay) ``` ``` ## [1] -2 ``` --- ## Measures of variation {.build} Standard deviation and Standard error (of the mean). ```r sd(flights_no_miss$dep_delay) ``` ``` ## [1] 40.21006 ``` ```r var(flights_no_miss$dep_delay) ``` ``` ## [1] 1616.849 ``` ```r se<-sd(flights_no_miss$dep_delay)/sqrt(length(flights$dep_delay)) se # standard error ``` ``` ## [1] 0.06928898 ``` --- ## 95% Confidence interval. ```r # 95 CI UL<- (mean(flights_no_miss$dep_delay) + 1.96*se) LL<- (mean(flights_no_miss$dep_delay) - 1.96*se) UL ``` ``` ## [1] 12.77488 ``` ```r LL ``` ``` ## [1] 12.50326 ``` <img src="CI.jpg" width="150px" /> --- ##Five number summary. minimum, first quartile (Q1), median, third quartile (Q3), maximum. ```r fivenum(flights_no_miss$dep_delay) ``` ``` ## [1] -43 -5 -2 11 1301 ``` ```r summary(flights_no_miss$dep_delay) ``` ``` ## Min. 1st Qu. Median Mean 3rd Qu. Max. ## -43.00 -5.00 -2.00 12.64 11.00 1301.00 ``` --- ## Interquartile range IQR: Q3 - Q1. Another measure of variation. ```r IQR(flights_no_miss$dep_delay) ``` ``` ## [1] 16 ``` --- ## Boxplot ```r boxplot(flights_no_miss$dep_delay) ``` 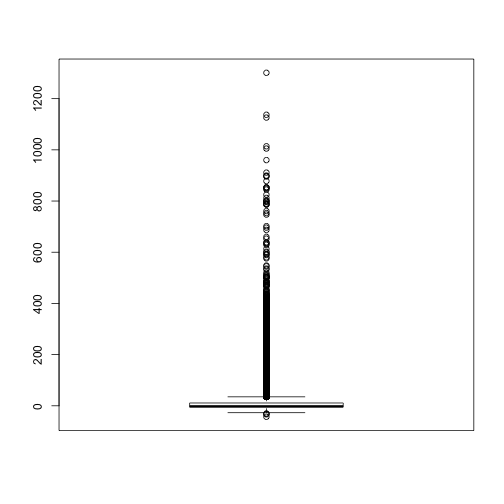<!-- --> --- ## First useful thing? Now use a package to make a PRISMA flow chart. --- ## PRISMA flow chart in R. ```r library(PRISMAstatement) prisma(found = 750, found_other = 123, no_dupes = 776, screened = 776, screen_exclusions = 13, full_text = 763, full_text_exclusions = 17, qualitative = 746, quantitative = 319, width = 800, height = 800) ``` --- ## Output chart <div class="figure" style="text-align: center"> <div id="htmlwidget-975252a1a3e5956b2825" style="width:350px;height:504px;" class="grViz html-widget"></div> <script type="application/json" data-for="htmlwidget-975252a1a3e5956b2825">{"x":{"diagram":"digraph prisma {\n node [shape=\"box\", fontsize = 10];\n graph [splines=ortho, nodesep=1, dpi = 72]\n a -> nodups;\n b -> nodups;\n a [label=\"Records identified through\ndatabase searching\n(n = 750)\"];\n b [label=\"Additional records identified\nthrough other sources\n(n = 123)\"]\n nodups -> incex;\n nodups [label=\"Records after duplicates removed\n(n = 776)\"];\n incex -> {ex; ft}\n incex [label=\"Records screened\n(n = 776)\"];\n ex [label=\"Records excluded\n(n = 13)\"];\n {rank=same; incex ex}\n ft -> {qual; ftex};\n ft [label=\"Full-text articles assessed\nfor eligibility\n(n = 763)\"];\n {rank=same; ft ftex}\n ftex [label=\"Full-text articles excluded,\nwith reasons\n(n = 17)\"];\n qual -> quant\n qual [label=\"Studies included in qualitative synthesis\n(n = 746)\"];\n quant [label=\"Studies included in\nquantitative synthesis\n(meta-analysis)\n(n = 319)\"];\n }","config":{"engine":"dot","options":null}},"evals":[],"jsHooks":[]}</script> <p class="caption">PRISMA Flow chart</p> </div> --- ## Exercise... . Load the flights dataset. Calculate the mean delay in arrival for Delta Airlines (DL) (use filter()) Calculate the associated 95% confidence interval. Do the same for United Airlines (UA) and compare the two. Do their confidence intervals overlap? Calculate the mode for the delay in arrival for at JFK airport. save a dataset as .sav with only departing flights from JFK airport. --- ## Any Questions? [http://tvpollet.github.io](http://tvpollet.github.io) Twitter: @tvpollet <img src="https://media.giphy.com/media/3ohzdRoOp1FUYbtGDu/giphy.gif" width="600px" style="display: block; margin: auto;" /> --- ## Acknowledgments * Numerous students and colleagues. Any mistakes are my own. * My colleagues who helped me with regards to meta-analysis specifically: Nexhmedin Morina, Stijn Peperkoorn, Gert Stulp, Mirre Simons, Johannes Honekopp. * [HBES](www.hbes.com) and [LECH](https://www.lechufrn.com/) for funding this workshop. Those who have funded me (not these studies per se): [NWO](www.nwo.nl), [Templeton](www.templeton.org), [NIAS](http://nias.knaw.nl). * You for listening! <img src="https://media.giphy.com/media/10avZ0rqdGFyfu/giphy.gif" width="300px" style="display: block; margin: auto;" /> --- ## References and further reading (errors = blame RefManageR) <p><cite>Aert, R. C. M. van, J. M. Wicherts, and M. A. L. M. van Assen (2016). “Conducting Meta-Analyses Based on p Values: Reservations and Recommendations for Applying p-Uniform and p-Curve”. In: <em>Perspectives on Psychological Science</em> 11.5, pp. 713-729. DOI: <a href="https://doi.org/10.1177/1745691616650874">10.1177/1745691616650874</a>. eprint: https://doi.org/10.1177/1745691616650874.</cite></p> <p><cite>Aloe, A. M. and C. G. Thompson (2013). “The Synthesis of Partial Effect Sizes”. In: <em>Journal of the Society for Social Work and Research</em> 4.4, pp. 390-405. DOI: <a href="https://doi.org/10.5243/jsswr.2013.24">10.5243/jsswr.2013.24</a>. eprint: https://doi.org/10.5243/jsswr.2013.24.</cite></p> <p><cite>Assink, M. and C. J. Wibbelink (2016). “Fitting Three-Level Meta-Analytic Models in R: A Step-by-Step Tutorial”. In: <em>The Quantitative Methods for Psychology</em> 12.3, pp. 154-174. ISSN: 2292-1354.</cite></p> <p><cite>Barendregt, J. J, S. A. Doi, Y. Y. Lee, et al. (2013). “Meta-Analysis of Prevalence”. In: <em>Journal of Epidemiology and Community Health</em> 67.11, pp. 974-978. ISSN: 0143-005X. DOI: <a href="https://doi.org/10.1136/jech-2013-203104">10.1136/jech-2013-203104</a>.</cite></p> <p><cite>Becker, B. J. and M. Wu (2007). “The Synthesis of Regression Slopes in Meta-Analysis”. In: <em>Statistical science</em> 22.3, pp. 414-429. ISSN: 0883-4237.</cite></p> --- ## More refs 1. <p><cite>Borenstein, M, L. V. Hedges, J. P. Higgins, et al. (2009). <em>Introduction to Meta-Analysis</em>. John Wiley & Sons. ISBN: 1-119-96437-7.</cite></p> <p><cite>Burnham, K. P. and D. R. Anderson (2002). <em>Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach</em>. New York, NY: Springer. ISBN: 0-387-95364-7.</cite></p> <p><cite>Burnham, K. P. and D. R. Anderson (2004). “Multimodel Inference: Understanding AIC and BIC in Model Selection”. In: <em>Sociological Methods & Research</em> 33.2, pp. 261-304. ISSN: 0049-1241. DOI: <a href="https://doi.org/10.1177/0049124104268644">10.1177/0049124104268644</a>.</cite></p> <p><cite>Carter, E. C, F. D. Schönbrodt, W. M. Gervais, et al. (2019). “Correcting for Bias in Psychology: A Comparison of Meta-Analytic Methods”. In: <em>Advances in Methods and Practices in Psychological Science</em> 2.2, pp. 115-144. DOI: <a href="https://doi.org/10.1177/2515245919847196">10.1177/2515245919847196</a>.</cite></p> <p><cite>Chen, D. D. and K. E. Peace (2013). <em>Applied Meta-Analysis with R</em>. Chapman and Hall/CRC. ISBN: 1-4665-0600-8.</cite></p> --- ## More refs 2. <p><cite>Cheung, M. W. (2015a). “metaSEM: An R Package for Meta-Analysis Using Structural Equation Modeling”. In: <em>Frontiers in Psychology</em> 5, p. 1521. ISSN: 1664-1078. DOI: <a href="https://doi.org/10.3389/fpsyg.2014.01521">10.3389/fpsyg.2014.01521</a>.</cite></p> <p><cite>Cheung, M. W. (2015b). <em>Meta-Analysis: A Structural Equation Modeling Approach</em>. New York, NY: John Wiley & Sons. ISBN: 1-119-99343-1.</cite></p> <p><cite>Cooper, H. (2010). <em>Research Synthesis and Meta-Analysis: A Step-by-Step Approach</em>. 4th. Sage publications. ISBN: 1-4833-4704-4.</cite></p> <p><cite>Cooper, H, L. V. Hedges, and J. C. Valentine (2009). <em>The Handbook of Research Synthesis and Meta-Analysis</em>. New York: Russell Sage Foundation. ISBN: 1-61044-138-9.</cite></p> <p><cite>Cooper, H. and E. A. Patall (2009). “The Relative Benefits of Meta-Analysis Conducted with Individual Participant Data versus Aggregated Data.” In: <em>Psychological Methods</em> 14.2, pp. 165-176. ISSN: 1433806886. DOI: <a href="https://doi.org/10.1037/a0015565">10.1037/a0015565</a>.</cite></p> --- ## More refs 3. <p><cite>Crawley, M. J. (2013). <em>The R Book: Second Edition</em>. New York, NY: John Wiley & Sons. ISBN: 1-118-44896-0.</cite></p> <p><cite>Cumming, G. (2014). “The New Statistics”. In: <em>Psychological Science</em> 25.1, pp. 7-29. ISSN: 0956-7976. DOI: <a href="https://doi.org/10.1177/0956797613504966">10.1177/0956797613504966</a>.</cite></p> <p><cite>Dickersin, K. (2005). “Publication Bias: Recognizing the Problem, Understanding Its Origins and Scope, and Preventing Harm”. In: <em>Publication Bias in Meta-Analysis Prevention, Assessment and Adjustments</em>. Ed. by H. R. Rothstein, A. J. Sutton and M. Borenstein. Chichester, UK: John Wiley.</cite></p> <p><cite>Fisher, R. A. (1946). <em>Statistical Methods for Research Workers</em>. 10th ed. Edinburgh, UK: Oliver and Boyd.</cite></p> <p><cite>Flore, P. C. and J. M. Wicherts (2015). “Does Stereotype Threat Influence Performance of Girls in Stereotyped Domains? A Meta-Analysis”. In: <em>Journal of School Psychology</em> 53.1, pp. 25-44. ISSN: 0022-4405. DOI: <a href="https://doi.org/10.1016/j.jsp.2014.10.002">10.1016/j.jsp.2014.10.002</a>.</cite></p> --- ## More refs 4. <p><cite>Galbraith, R. F. (1994). “Some Applications of Radial Plots”. In: <em>Journal of the American Statistical Association</em> 89.428, pp. 1232-1242. ISSN: 0162-1459. DOI: <a href="https://doi.org/10.1080/01621459.1994.10476864">10.1080/01621459.1994.10476864</a>.</cite></p> <p><cite>Glass, G. V. (1976). “Primary, Secondary, and Meta-Analysis of Research”. In: <em>Educational researcher</em> 5.10, pp. 3-8. ISSN: 0013-189X. DOI: <a href="https://doi.org/10.3102/0013189X005010003">10.3102/0013189X005010003</a>.</cite></p> <p><cite>Goh, J. X, J. A. Hall, and R. Rosenthal (2016). “Mini Meta-Analysis of Your Own Studies: Some Arguments on Why and a Primer on How”. In: <em>Social and Personality Psychology Compass</em> 10.10, pp. 535-549. ISSN: 1751-9004. DOI: <a href="https://doi.org/10.1111/spc3.12267">10.1111/spc3.12267</a>.</cite></p> <p><cite>Harrell, F. E. (2015). <em>Regression Modeling Strategies</em>. 2nd. Springer Series in Statistics. New York, NY: Springer New York. ISBN: 978-1-4419-2918-1. DOI: <a href="https://doi.org/10.1007/978-1-4757-3462-1">10.1007/978-1-4757-3462-1</a>.</cite></p> <p><cite>Harrer, M., P. Cuijpers, and D. D. Ebert (2019). <em>Doing Meta-Analysis in R: A Hands-on Guide</em>. https://bookdown.org/MathiasHarrer/Doing\_ Meta\_ Analysis\_ in\_ R/.</cite></p> --- ## More refs 5. <p><cite>Hartung, J. and G. Knapp (2001). “On Tests of the Overall Treatment Effect in Meta-Analysis with Normally Distributed Responses”. In: <em>Statistics in Medicine</em> 20.12, pp. 1771-1782. DOI: <a href="https://doi.org/10.1002/sim.791">10.1002/sim.791</a>.</cite></p> <p><cite>Hayes, A. F. and K. Krippendorff (2007). “Answering the Call for a Standard Reliability Measure for Coding Data”. In: <em>Communication Methods and Measures</em> 1.1, pp. 77-89. ISSN: 1931-2458. DOI: <a href="https://doi.org/10.1080/19312450709336664">10.1080/19312450709336664</a>.</cite></p> <p><cite>Hedges, L. V. (1981). “Distribution Theory for Glass's Estimator of Effect Size and Related Estimators”. In: <em>Journal of Educational Statistics</em> 6.2, pp. 107-128. DOI: <a href="https://doi.org/10.3102/10769986006002107">10.3102/10769986006002107</a>.</cite></p> <p><cite>Hedges, L. V. (1984). “Estimation of Effect Size under Nonrandom Sampling: The Effects of Censoring Studies Yielding Statistically Insignificant Mean Differences”. In: <em>Journal of Educational Statistics</em> 9.1, pp. 61-85. ISSN: 0362-9791. DOI: <a href="https://doi.org/10.3102/10769986009001061">10.3102/10769986009001061</a>.</cite></p> <p><cite>Hedges, L. V. and I. Olkin (1980). “Vote-Counting Methods in Research Synthesis.” In: <em>Psychological bulletin</em> 88.2, pp. 359-369. ISSN: 1939-1455. DOI: <a href="https://doi.org/10.1037/0033-2909.88.2.359">10.1037/0033-2909.88.2.359</a>.</cite></p> --- ## More refs 6. <p><cite>Higgins, J. P. T. and S. G. Thompson (2002). “Quantifying Heterogeneity in a Meta-Analysis”. In: <em>Statistics in Medicine</em> 21.11, pp. 1539-1558. DOI: <a href="https://doi.org/10.1002/sim.1186">10.1002/sim.1186</a>.</cite></p> <p><cite>Higgins, J. P. T, S. G. Thompson, J. J. Deeks, et al. (2003). “Measuring Inconsistency in Meta-Analyses”. In: <em>BMJ</em> 327.7414, pp. 557-560. ISSN: 0959-8138. DOI: <a href="https://doi.org/10.1136/bmj.327.7414.557">10.1136/bmj.327.7414.557</a>.</cite></p> <p><cite>Higgins, J, S. Thompson, J. Deeks, et al. (2002). “Statistical Heterogeneity in Systematic Reviews of Clinical Trials: A Critical Appraisal of Guidelines and Practice”. In: <em>Journal of Health Services Research & Policy</em> 7.1, pp. 51-61. DOI: <a href="https://doi.org/10.1258/1355819021927674">10.1258/1355819021927674</a>.</cite></p> <p><cite>Hirschenhauser, K. and R. F. Oliveira (2006). “Social Modulation of Androgens in Male Vertebrates: Meta-Analyses of the Challenge Hypothesis”. In: <em>Animal Behaviour</em> 71.2, pp. 265-277. ISSN: 0003-3472. DOI: <a href="https://doi.org/10.1016/j.anbehav.2005.04.014">10.1016/j.anbehav.2005.04.014</a>.</cite></p> <p><cite>Ioannidis, J. P. (2008). “Why Most Discovered True Associations Are Inflated”. In: <em>Epidemiology</em> 19.5, pp. 640-648. ISSN: 1044-3983.</cite></p> --- ## More refs 7. <p><cite>Jackson, D, M. Law, G. Rücker, et al. (2017). “The Hartung-Knapp Modification for Random-Effects Meta-Analysis: A Useful Refinement but Are There Any Residual Concerns?” In: <em>Statistics in Medicine</em> 36.25, pp. 3923-3934. DOI: <a href="https://doi.org/10.1002/sim.7411">10.1002/sim.7411</a>. eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/sim.7411.</cite></p> <p><cite>Jacobs, P. and W. Viechtbauer (2016). “Estimation of the Biserial Correlation and Its Sampling Variance for Use in Meta-Analysis”. In: <em>Research Synthesis Methods</em> 8.2, pp. 161-180. DOI: <a href="https://doi.org/10.1002/jrsm.1218">10.1002/jrsm.1218</a>.</cite></p> <p><cite>Koricheva, J, J. Gurevitch, and K. Mengersen (2013). <em>Handbook of Meta-Analysis in Ecology and Evolution</em>. Princeton, NJ: Princeton University Press. ISBN: 0-691-13729-3.</cite></p> <p><cite>Kovalchik, S. (2013). <em>Tutorial On Meta-Analysis In R - R useR! Conference 2013</em>.</cite></p> <p><cite>Lipsey, M. W. and D. B. Wilson (2001). <em>Practical Meta-Analysis.</em> London: SAGE publications, Inc. ISBN: 0-7619-2167-2.</cite></p> --- ## More refs 8. <p><cite>Littell, J. H, J. Corcoran, and V. Pillai (2008). <em>Systematic Reviews and Meta-Analysis</em>. Oxford, UK: Oxford University Press. ISBN: 0-19-532654-7.</cite></p> <p><cite>McShane, B. B, U. Böckenholt, and K. T. Hansen (2016). “Adjusting for Publication Bias in Meta-Analysis: An Evaluation of Selection Methods and Some Cautionary Notes”. In: <em>Perspectives on Psychological Science</em> 11.5, pp. 730-749. DOI: <a href="https://doi.org/10.1177/1745691616662243">10.1177/1745691616662243</a>. eprint: https://doi.org/10.1177/1745691616662243.</cite></p> <p><cite>Mengersen, K, C. Schmidt, M. Jennions, et al. (2013). “Statistical Models and Approaches to Inference”. In: <em>Handbook of Meta-Analysis in Ecology and Evolution</em>. Ed. by Koricheva, J, J. Gurevitch and Mengersen, Kerrie. Princeton, NJ: Princeton University Press, pp. 89-107.</cite></p> <p><cite>Methley, A. M, S. Campbell, C. Chew-Graham, et al. (2014). “PICO, PICOS and SPIDER: A Comparison Study of Specificity and Sensitivity in Three Search Tools for Qualitative Systematic Reviews”. Eng. In: <em>BMC health services research</em> 14, pp. 579-579. ISSN: 1472-6963. DOI: <a href="https://doi.org/10.1186/s12913-014-0579-0">10.1186/s12913-014-0579-0</a>.</cite></p> <p><cite>Morina, N, K. Stam, T. V. Pollet, et al. (2018). “Prevalence of Depression and Posttraumatic Stress Disorder in Adult Civilian Survivors of War Who Stay in War-Afflicted Regions. A Systematic Review and Meta-Analysis of Epidemiological Studies”. In: <em>Journal of Affective Disorders</em> 239, pp. 328-338. ISSN: 0165-0327. DOI: <a href="https://doi.org/10.1016/j.jad.2018.07.027">10.1016/j.jad.2018.07.027</a>.</cite></p> --- ## More refs 9. <p><cite>Nakagawa, S, D. W. A. Noble, A. M. Senior, et al. (2017). “Meta-Evaluation of Meta-Analysis: Ten Appraisal Questions for Biologists”. In: <em>BMC Biology</em> 15.1, p. 18. ISSN: 1741-7007. DOI: <a href="https://doi.org/10.1186/s12915-017-0357-7">10.1186/s12915-017-0357-7</a>.</cite></p> <p><cite>Pastor, D. A. and R. A. Lazowski (2018). “On the Multilevel Nature of Meta-Analysis: A Tutorial, Comparison of Software Programs, and Discussion of Analytic Choices”. In: <em>Multivariate Behavioral Research</em> 53.1, pp. 74-89. DOI: <a href="https://doi.org/10.1080/00273171.2017.1365684">10.1080/00273171.2017.1365684</a>.</cite></p> <p><cite>Poole, C. and S. Greenland (1999). “Random-Effects Meta-Analyses Are Not Always Conservative”. In: <em>American Journal of Epidemiology</em> 150.5, pp. 469-475. ISSN: 0002-9262. DOI: <a href="https://doi.org/10.1093/oxfordjournals.aje.a010035">10.1093/oxfordjournals.aje.a010035</a>. eprint: http://oup.prod.sis.lan/aje/article-pdf/150/5/469/286690/150-5-469.pdf.</cite></p> <p><cite>Popper, K. (1959). <em>The Logic of Scientific Discovery</em>. London, UK: Hutchinson. ISBN: 1-134-47002-9.</cite></p> <p><cite>Roberts, P. D, G. B. Stewart, and A. S. Pullin (2006). “Are Review Articles a Reliable Source of Evidence to Support Conservation and Environmental Management? A Comparison with Medicine”. In: <em>Biological conservation</em> 132.4, pp. 409-423. ISSN: 0006-3207.</cite></p> --- ## More refs 10. <p><cite>Rosenberg, M. S, H. R. Rothstein, and J. Gurevitch (2013). “Effect Sizes: Conventional Choices and Calculations”. In: <em>Handbook of Meta-analysis in Ecology and Evolution</em>, pp. 61-71.</cite></p> <p><cite>Röver, C, G. Knapp, and T. Friede (2015). “Hartung-Knapp-Sidik-Jonkman Approach and Its Modification for Random-Effects Meta-Analysis with Few Studies”. In: <em>BMC Medical Research Methodology</em> 15.1, p. 99. ISSN: 1471-2288. DOI: <a href="https://doi.org/10.1186/s12874-015-0091-1">10.1186/s12874-015-0091-1</a>.</cite></p> <p><cite>Schwarzer, G, J. R. Carpenter, and G. Rücker (2015). <em>Meta-Analysis with R</em>. New York, NY: Springer. ISBN: 3-319-21415-2.</cite></p> <p><cite>Schwarzer, G, H. Chemaitelly, L. J. Abu-Raddad, et al. “Seriously Misleading Results Using Inverse of Freeman-Tukey Double Arcsine Transformation in Meta-Analysis of Single Proportions”. In: <em>Research Synthesis Methods</em> 0.0. DOI: <a href="https://doi.org/10.1002/jrsm.1348">10.1002/jrsm.1348</a>. eprint: https://onlinelibrary.wiley.com/doi/pdf/10.1002/jrsm.1348.</cite></p> <p><cite>Simmons, J. P, L. D. Nelson, and U. Simonsohn (2011). “False-Positive Psychology”. In: <em>Psychological Science</em> 22.11, pp. 1359-1366. ISSN: 0956-7976. DOI: <a href="https://doi.org/10.1177/0956797611417632">10.1177/0956797611417632</a>.</cite></p> --- ## More refs 11. <p><cite>Simonsohn, U, L. D. Nelson, and J. P. Simmons (2014). “P-Curve: A Key to the File-Drawer.” In: <em>Journal of Experimental Psychology: General</em> 143.2, pp. 534-547. ISSN: 1939-2222. DOI: <a href="https://doi.org/10.1037/a0033242">10.1037/a0033242</a>.</cite></p> <p><cite>Sterne, J. A. C, A. J. Sutton, J. P. A. Ioannidis, et al. (2011). “Recommendations for Examining and Interpreting Funnel Plot Asymmetry in Meta-Analyses of Randomised Controlled Trials”. In: <em>BMJ</em> 343.jul22 1, pp. d4002-d4002. ISSN: 0959-8138. DOI: <a href="https://doi.org/10.1136/bmj.d4002">10.1136/bmj.d4002</a>.</cite></p> <p><cite>Veroniki, A. A, D. Jackson, W. Viechtbauer, et al. (2016). “Methods to Estimate the Between-Study Variance and Its Uncertainty in Meta-Analysis”. Eng. In: <em>Research synthesis methods</em> 7.1, pp. 55-79. ISSN: 1759-2887. DOI: <a href="https://doi.org/10.1002/jrsm.1164">10.1002/jrsm.1164</a>.</cite></p> <p><cite>Viechtbauer, W. (2015). “Package ‘metafor’: Meta-Analysis Package for R”. </p></cite></p> <p><cite>Weiss, B. and J. Daikeler (2017). <em>Syllabus for Course: “Meta-Analysis in Survey Methodology", 6th Summer Workshop (GESIS)</em>.</cite></p> --- ## More refs 12. <p><cite>Wickham, H. and G. Grolemund (2016). <em>R for Data Science</em>. Sebastopol, CA: O'Reilly..</cite></p> <p><cite>Wiernik, B. (2015). <em>A Brief Introduction to Meta-Analysis</em>.</cite></p> <p><cite>Wiksten, A, G. Rücker, and G. Schwarzer (2016). “Hartung-Knapp Method Is Not Always Conservative Compared with Fixed-Effect Meta-Analysis”. In: <em>Statistics in Medicine</em> 35.15, pp. 2503-2515. DOI: <a href="https://doi.org/10.1002/sim.6879">10.1002/sim.6879</a>.</cite></p> <p><cite>Wingfield, J. C, R. E. Hegner, A. M. Dufty Jr, et al. (1990). “The" Challenge Hypothesis": Theoretical Implications for Patterns of Testosterone Secretion, Mating Systems, and Breeding Strategies”. In: <em>American Naturalist</em> 136, pp. 829-846. ISSN: 0003-0147.</cite></p> <p><cite>Yeaton, W. H. and P. M. Wortman (1993). “On the Reliability of Meta-Analytic Reviews: The Role of Intercoder Agreement”. In: <em>Evaluation Review</em> 17.3, pp. 292-309. ISSN: 0193-841X. DOI: <a href="https://doi.org/10.1177/0193841X9301700303">10.1177/0193841X9301700303</a>.</cite></p> --- ## More refs 13. --- ## More refs 14.